The amphipod crustacean Parhyale hawaiensis has germband formation that is different from the long-germ type of Drosophila.
Germband formation in Parhyale is sequential like in short-germ insects.
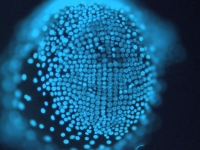
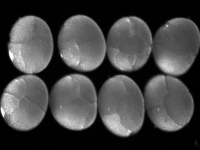
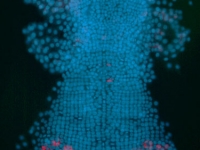
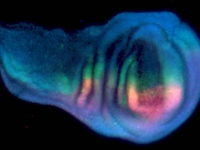
 A day old Parhyale embryo that was injected at the 1-cell stage with supercoiled plasmid construct pECE-DsRed-NLS. This construct consists of a ~9kb promoter/enhancer fragment from an endogenous Ef1-alpha gene that constitutively drives expression of a nuclear localized variant of DsRed fluorescent protein. In fact, expression appears to be sub-nuclear, for unknown reasons. The nuclei of all cells are fluorescing. Fluorescence first appears at around 18 hours, about the time of gastrulation. We believe that this coincides with the onset of zygotic tranion. In the pictured embryo a germdisc has formed at the anterior end of the egg (top). A fluorescent image has been overlaid on a brightfield image.
A day old Parhyale embryo that was injected at the 1-cell stage with supercoiled plasmid construct pECE-DsRed-NLS. This construct consists of a ~9kb promoter/enhancer fragment from an endogenous Ef1-alpha gene that constitutively drives expression of a nuclear localized variant of DsRed fluorescent protein. In fact, expression appears to be sub-nuclear, for unknown reasons. The nuclei of all cells are fluorescing. Fluorescence first appears at around 18 hours, about the time of gastrulation. We believe that this coincides with the onset of zygotic tranion. In the pictured embryo a germdisc has formed at the anterior end of the egg (top). A fluorescent image has been overlaid on a brightfield image.
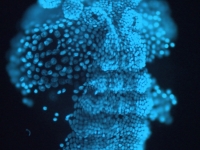
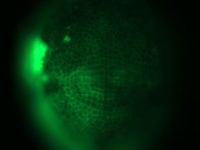
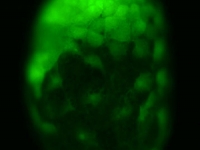 Fluorescent image of a day old Parhyale embryo that was injected at the 1-cell stage with supercoiled plasmid construct pECE-Timer. In this case, the ef1alpha promoter/enhancer drives cytoplasmic expression of Timer, a DsRed variant that fluoresces green for a few hours before the protein folds into its mature conformation, at which point it fluoresces red. In the pictured embryo a germdisc has formed at the anterior end of the egg (top).
Courtesy of E. Jay Rehm
Fluorescent image of a day old Parhyale embryo that was injected at the 1-cell stage with supercoiled plasmid construct pECE-Timer. In this case, the ef1alpha promoter/enhancer drives cytoplasmic expression of Timer, a DsRed variant that fluoresces green for a few hours before the protein folds into its mature conformation, at which point it fluoresces red. In the pictured embryo a germdisc has formed at the anterior end of the egg (top).
Courtesy of E. Jay Rehm
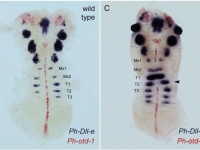
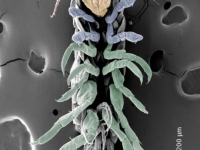
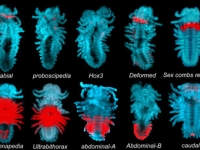
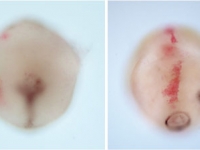
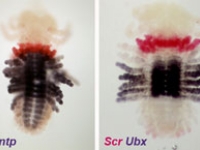
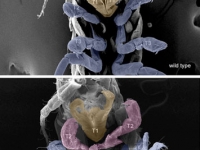 Ventral view SEM comparing a wild-type Parhyale hatchling (top) with a hatchling that had been injected with siRNAs targeting the Parhyale ortholog of the Hox gene Ultrabithorax (PhUbx) (bottom). Appendages are highlighted according to identity (maxillipeds on T1 are orange, walking legs are blue, transformed appendages on T2 with ectopic branches and a partial maxilliped-like identity are pink).
Courtesy of Danielle Liubicich
Ventral view SEM comparing a wild-type Parhyale hatchling (top) with a hatchling that had been injected with siRNAs targeting the Parhyale ortholog of the Hox gene Ultrabithorax (PhUbx) (bottom). Appendages are highlighted according to identity (maxillipeds on T1 are orange, walking legs are blue, transformed appendages on T2 with ectopic branches and a partial maxilliped-like identity are pink).
Courtesy of Danielle Liubicich
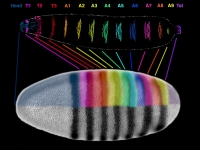
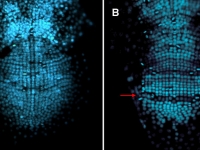
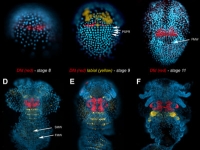
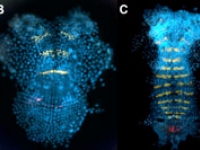
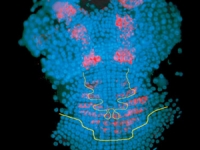 Parhyale odd-paired (opa-1) expression in the germband (anterior is up). The darker yellow line traces the first of two highly organized rounds of mitotic division, which begin medially before spreading laterally as a wavefront. Below this lie transverse rows of parasegment precursor cells, each of which represents a future parasegment. The lighter yellow line indicates the second mitotic wavefront, which results in a parasegment containing 4 cell rows. As is clear from the false-colored in situ (red), opa-1 expression begins shortly after the first mitotic division. It is expressed only in the more posterior row of daughter cells following the first mitotic event. Anterior to the germband, opa-1 is expressed in appendage fields of the head parasegments, which are further along in development. (In situ image courtesy of Ron Parchem.)
Courtesy of E. Jay Rehm
Parhyale odd-paired (opa-1) expression in the germband (anterior is up). The darker yellow line traces the first of two highly organized rounds of mitotic division, which begin medially before spreading laterally as a wavefront. Below this lie transverse rows of parasegment precursor cells, each of which represents a future parasegment. The lighter yellow line indicates the second mitotic wavefront, which results in a parasegment containing 4 cell rows. As is clear from the false-colored in situ (red), opa-1 expression begins shortly after the first mitotic division. It is expressed only in the more posterior row of daughter cells following the first mitotic event. Anterior to the germband, opa-1 is expressed in appendage fields of the head parasegments, which are further along in development. (In situ image courtesy of Ron Parchem.)
Courtesy of E. Jay Rehm
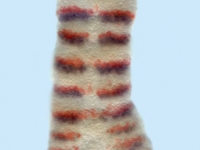
 Expression of hedgehog (hh) in an ablated Parhyale hawaiensis embryo. (Ventral view of posterior ectoderm, anterior is up and posterior is down.) The right side of the embryo shows wild-type hh expression (white arrowheads) where hh is expressed in every fourth cell row. In the left side of the embryo, all non-hh expressing cells have been ablated in two adjacent parasegments. This manipulation prevents at least one hh-expressing cell from coming in contact with any cell besides other hh-expressing cells (yellow arrowhead). The manipulation does not lead to a decrease in hh expression, suggesting that hh expression in these cells is a cell-intrinsic property.
Courtesy of Mario Vargas-Vila
Expression of hedgehog (hh) in an ablated Parhyale hawaiensis embryo. (Ventral view of posterior ectoderm, anterior is up and posterior is down.) The right side of the embryo shows wild-type hh expression (white arrowheads) where hh is expressed in every fourth cell row. In the left side of the embryo, all non-hh expressing cells have been ablated in two adjacent parasegments. This manipulation prevents at least one hh-expressing cell from coming in contact with any cell besides other hh-expressing cells (yellow arrowhead). The manipulation does not lead to a decrease in hh expression, suggesting that hh expression in these cells is a cell-intrinsic property.
Courtesy of Mario Vargas-Vila
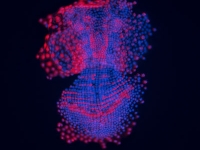 Expression of Phhes-1, an ortholog of the Drosophila hairy/enhancer of split, in Parhyale hawaiiensis. This is one of a set of genes that are expressed in a row-specific manner after each wave of mitotic division in the Parhyale ectoderm, and can serve as a marker of cell fate in experiments such as those using the Notch pathway inhibitor DAPT.
Courtesy of Mike Perry
Expression of Phhes-1, an ortholog of the Drosophila hairy/enhancer of split, in Parhyale hawaiiensis. This is one of a set of genes that are expressed in a row-specific manner after each wave of mitotic division in the Parhyale ectoderm, and can serve as a marker of cell fate in experiments such as those using the Notch pathway inhibitor DAPT.
Courtesy of Mike Perry
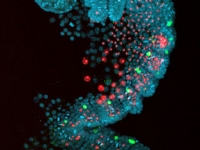
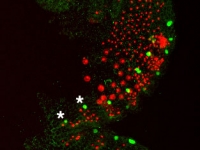 Even-skipped2 protein (green) is expressed in a subset of the segmental mesoderm (red). The embryo in this set of photos was injected with DsRed-NLS mRNA into one of the mesodermal precursor cells at the eight-cell stage (7hrs of development). It was fixed about four days later and stained with an antibody to DsRed-NLS protein, in red, and with an antibody to Ph-Eve2, in green. All the nuclei of the embryo are labeled with Draq5, in blue. The first image is a merge of all three stains while the second only shows Ph-Eve2 and segmental mesoderm staining. The embryo is orientated on its side, ventral to the right, and anterior to the top. Developing antennae are near the top, and limb buds are along the right side of the embryo. Ph-Eve2 staining is seen in the anterior mesoderm of each segment, marked by a nearby * in two of the younger segments. Ph-Eve2 is also seen in the youngest segmental mesoderm and ectoderm, the most posterior cells of the embryo, and in the nervous system, the cells along the ventral edge of the embryo.
Courtesy of Roberta Hannibal
Even-skipped2 protein (green) is expressed in a subset of the segmental mesoderm (red). The embryo in this set of photos was injected with DsRed-NLS mRNA into one of the mesodermal precursor cells at the eight-cell stage (7hrs of development). It was fixed about four days later and stained with an antibody to DsRed-NLS protein, in red, and with an antibody to Ph-Eve2, in green. All the nuclei of the embryo are labeled with Draq5, in blue. The first image is a merge of all three stains while the second only shows Ph-Eve2 and segmental mesoderm staining. The embryo is orientated on its side, ventral to the right, and anterior to the top. Developing antennae are near the top, and limb buds are along the right side of the embryo. Ph-Eve2 staining is seen in the anterior mesoderm of each segment, marked by a nearby * in two of the younger segments. Ph-Eve2 is also seen in the youngest segmental mesoderm and ectoderm, the most posterior cells of the embryo, and in the nervous system, the cells along the ventral edge of the embryo.
Courtesy of Roberta Hannibal
 Chirality in snails. a, Two gastropod species with different chirality: a sinistral specimen of Busycon pulleyi (Melongenidae) from Texas, USA (left) and a dextral specimen of Fusinus salisbury (Fasciolariidae) from Palawan, Philippines (right). b, Sinistral (left) and dextral (right) shells of Amphidromus perversus from Madura Island, Indonesia.
Courtesy of Cristina Grande
Chirality in snails. a, Two gastropod species with different chirality: a sinistral specimen of Busycon pulleyi (Melongenidae) from Texas, USA (left) and a dextral specimen of Fusinus salisbury (Fasciolariidae) from Palawan, Philippines (right). b, Sinistral (left) and dextral (right) shells of Amphidromus perversus from Madura Island, Indonesia.
Courtesy of Cristina Grande
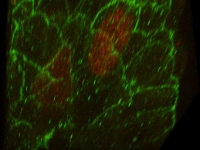
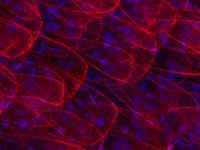
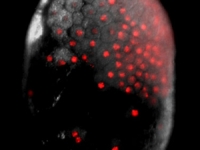
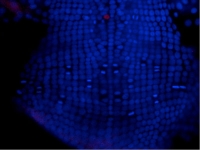 A Parhyale embryo stained for Prospero (red) and DNA (blue). Prospero expression initiates in the d1h cell lineage just after the first differential division. This is the homologous neuroblast that in Diastylis has been observed to give rise to both neural and ectodermally fated cells.
Courtesy of Paul Liu
A Parhyale embryo stained for Prospero (red) and DNA (blue). Prospero expression initiates in the d1h cell lineage just after the first differential division. This is the homologous neuroblast that in Diastylis has been observed to give rise to both neural and ectodermally fated cells.
Courtesy of Paul Liu
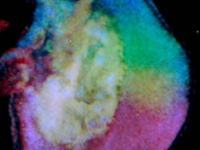
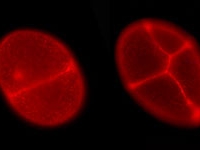
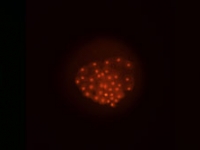 A flattened P.hawaiensis embryo at germ cap stage (~25h of development). One cell of this embryo was previously injected at the eight-cell stage with a nuclear localized mRNA that codes for a protein which fluoresces in both green (far left) and red (middle) when hit with blue or green light, respectively. The far right panel shows the two other panels in overlay. Embryo mounted by Mario Vargas-Vila.
Courtesy of Crystal Chaw
A flattened P.hawaiensis embryo at germ cap stage (~25h of development). One cell of this embryo was previously injected at the eight-cell stage with a nuclear localized mRNA that codes for a protein which fluoresces in both green (far left) and red (middle) when hit with blue or green light, respectively. The far right panel shows the two other panels in overlay. Embryo mounted by Mario Vargas-Vila.
Courtesy of Crystal Chaw
 A day old Parhyale embryo that was injected at the 1-cell stage with supercoiled plasmid construct pECE-DsRed-NLS. This construct consists of a ~9kb promoter/enhancer fragment from an endogenous Ef1-alpha gene that constitutively drives expression of a nuclear localized variant of DsRed fluorescent protein. In fact, expression appears to be sub-nuclear, for unknown reasons. The nuclei of all cells are fluorescing. Fluorescence first appears at around 18 hours, about the time of gastrulation. We believe that this coincides with the onset of zygotic tranion. In the pictured embryo a germdisc has formed at the anterior end of the egg (top). A fluorescent image has been overlaid on a brightfield image.
A day old Parhyale embryo that was injected at the 1-cell stage with supercoiled plasmid construct pECE-DsRed-NLS. This construct consists of a ~9kb promoter/enhancer fragment from an endogenous Ef1-alpha gene that constitutively drives expression of a nuclear localized variant of DsRed fluorescent protein. In fact, expression appears to be sub-nuclear, for unknown reasons. The nuclei of all cells are fluorescing. Fluorescence first appears at around 18 hours, about the time of gastrulation. We believe that this coincides with the onset of zygotic tranion. In the pictured embryo a germdisc has formed at the anterior end of the egg (top). A fluorescent image has been overlaid on a brightfield image.
 Fluorescent image of a day old Parhyale embryo that was injected at the 1-cell stage with supercoiled plasmid construct pECE-Timer. In this case, the ef1alpha promoter/enhancer drives cytoplasmic expression of Timer, a DsRed variant that fluoresces green for a few hours before the protein folds into its mature conformation, at which point it fluoresces red. In the pictured embryo a germdisc has formed at the anterior end of the egg (top).
Courtesy of E. Jay Rehm
Fluorescent image of a day old Parhyale embryo that was injected at the 1-cell stage with supercoiled plasmid construct pECE-Timer. In this case, the ef1alpha promoter/enhancer drives cytoplasmic expression of Timer, a DsRed variant that fluoresces green for a few hours before the protein folds into its mature conformation, at which point it fluoresces red. In the pictured embryo a germdisc has formed at the anterior end of the egg (top).
Courtesy of E. Jay Rehm
 Ventral view SEM comparing a wild-type Parhyale hatchling (top) with a hatchling that had been injected with siRNAs targeting the Parhyale ortholog of the Hox gene Ultrabithorax (PhUbx) (bottom). Appendages are highlighted according to identity (maxillipeds on T1 are orange, walking legs are blue, transformed appendages on T2 with ectopic branches and a partial maxilliped-like identity are pink).
Courtesy of Danielle Liubicich
Ventral view SEM comparing a wild-type Parhyale hatchling (top) with a hatchling that had been injected with siRNAs targeting the Parhyale ortholog of the Hox gene Ultrabithorax (PhUbx) (bottom). Appendages are highlighted according to identity (maxillipeds on T1 are orange, walking legs are blue, transformed appendages on T2 with ectopic branches and a partial maxilliped-like identity are pink).
Courtesy of Danielle Liubicich
 Parhyale odd-paired (opa-1) expression in the germband (anterior is up). The darker yellow line traces the first of two highly organized rounds of mitotic division, which begin medially before spreading laterally as a wavefront. Below this lie transverse rows of parasegment precursor cells, each of which represents a future parasegment. The lighter yellow line indicates the second mitotic wavefront, which results in a parasegment containing 4 cell rows. As is clear from the false-colored in situ (red), opa-1 expression begins shortly after the first mitotic division. It is expressed only in the more posterior row of daughter cells following the first mitotic event. Anterior to the germband, opa-1 is expressed in appendage fields of the head parasegments, which are further along in development. (In situ image courtesy of Ron Parchem.)
Courtesy of E. Jay Rehm
Parhyale odd-paired (opa-1) expression in the germband (anterior is up). The darker yellow line traces the first of two highly organized rounds of mitotic division, which begin medially before spreading laterally as a wavefront. Below this lie transverse rows of parasegment precursor cells, each of which represents a future parasegment. The lighter yellow line indicates the second mitotic wavefront, which results in a parasegment containing 4 cell rows. As is clear from the false-colored in situ (red), opa-1 expression begins shortly after the first mitotic division. It is expressed only in the more posterior row of daughter cells following the first mitotic event. Anterior to the germband, opa-1 is expressed in appendage fields of the head parasegments, which are further along in development. (In situ image courtesy of Ron Parchem.)
Courtesy of E. Jay Rehm
 Expression of hedgehog (hh) in an ablated Parhyale hawaiensis embryo. (Ventral view of posterior ectoderm, anterior is up and posterior is down.) The right side of the embryo shows wild-type hh expression (white arrowheads) where hh is expressed in every fourth cell row. In the left side of the embryo, all non-hh expressing cells have been ablated in two adjacent parasegments. This manipulation prevents at least one hh-expressing cell from coming in contact with any cell besides other hh-expressing cells (yellow arrowhead). The manipulation does not lead to a decrease in hh expression, suggesting that hh expression in these cells is a cell-intrinsic property.
Courtesy of Mario Vargas-Vila
Expression of hedgehog (hh) in an ablated Parhyale hawaiensis embryo. (Ventral view of posterior ectoderm, anterior is up and posterior is down.) The right side of the embryo shows wild-type hh expression (white arrowheads) where hh is expressed in every fourth cell row. In the left side of the embryo, all non-hh expressing cells have been ablated in two adjacent parasegments. This manipulation prevents at least one hh-expressing cell from coming in contact with any cell besides other hh-expressing cells (yellow arrowhead). The manipulation does not lead to a decrease in hh expression, suggesting that hh expression in these cells is a cell-intrinsic property.
Courtesy of Mario Vargas-Vila
 Expression of Phhes-1, an ortholog of the Drosophila hairy/enhancer of split, in Parhyale hawaiiensis. This is one of a set of genes that are expressed in a row-specific manner after each wave of mitotic division in the Parhyale ectoderm, and can serve as a marker of cell fate in experiments such as those using the Notch pathway inhibitor DAPT.
Courtesy of Mike Perry
Expression of Phhes-1, an ortholog of the Drosophila hairy/enhancer of split, in Parhyale hawaiiensis. This is one of a set of genes that are expressed in a row-specific manner after each wave of mitotic division in the Parhyale ectoderm, and can serve as a marker of cell fate in experiments such as those using the Notch pathway inhibitor DAPT.
Courtesy of Mike Perry
 Even-skipped2 protein (green) is expressed in a subset of the segmental mesoderm (red). The embryo in this set of photos was injected with DsRed-NLS mRNA into one of the mesodermal precursor cells at the eight-cell stage (7hrs of development). It was fixed about four days later and stained with an antibody to DsRed-NLS protein, in red, and with an antibody to Ph-Eve2, in green. All the nuclei of the embryo are labeled with Draq5, in blue. The first image is a merge of all three stains while the second only shows Ph-Eve2 and segmental mesoderm staining. The embryo is orientated on its side, ventral to the right, and anterior to the top. Developing antennae are near the top, and limb buds are along the right side of the embryo. Ph-Eve2 staining is seen in the anterior mesoderm of each segment, marked by a nearby * in two of the younger segments. Ph-Eve2 is also seen in the youngest segmental mesoderm and ectoderm, the most posterior cells of the embryo, and in the nervous system, the cells along the ventral edge of the embryo.
Courtesy of Roberta Hannibal
Even-skipped2 protein (green) is expressed in a subset of the segmental mesoderm (red). The embryo in this set of photos was injected with DsRed-NLS mRNA into one of the mesodermal precursor cells at the eight-cell stage (7hrs of development). It was fixed about four days later and stained with an antibody to DsRed-NLS protein, in red, and with an antibody to Ph-Eve2, in green. All the nuclei of the embryo are labeled with Draq5, in blue. The first image is a merge of all three stains while the second only shows Ph-Eve2 and segmental mesoderm staining. The embryo is orientated on its side, ventral to the right, and anterior to the top. Developing antennae are near the top, and limb buds are along the right side of the embryo. Ph-Eve2 staining is seen in the anterior mesoderm of each segment, marked by a nearby * in two of the younger segments. Ph-Eve2 is also seen in the youngest segmental mesoderm and ectoderm, the most posterior cells of the embryo, and in the nervous system, the cells along the ventral edge of the embryo.
Courtesy of Roberta Hannibal
 Chirality in snails. a, Two gastropod species with different chirality: a sinistral specimen of Busycon pulleyi (Melongenidae) from Texas, USA (left) and a dextral specimen of Fusinus salisbury (Fasciolariidae) from Palawan, Philippines (right). b, Sinistral (left) and dextral (right) shells of Amphidromus perversus from Madura Island, Indonesia.
Courtesy of Cristina Grande
Chirality in snails. a, Two gastropod species with different chirality: a sinistral specimen of Busycon pulleyi (Melongenidae) from Texas, USA (left) and a dextral specimen of Fusinus salisbury (Fasciolariidae) from Palawan, Philippines (right). b, Sinistral (left) and dextral (right) shells of Amphidromus perversus from Madura Island, Indonesia.
Courtesy of Cristina Grande
 A Parhyale embryo stained for Prospero (red) and DNA (blue). Prospero expression initiates in the d1h cell lineage just after the first differential division. This is the homologous neuroblast that in Diastylis has been observed to give rise to both neural and ectodermally fated cells.
Courtesy of Paul Liu
A Parhyale embryo stained for Prospero (red) and DNA (blue). Prospero expression initiates in the d1h cell lineage just after the first differential division. This is the homologous neuroblast that in Diastylis has been observed to give rise to both neural and ectodermally fated cells.
Courtesy of Paul Liu
 A flattened P.hawaiensis embryo at germ cap stage (~25h of development). One cell of this embryo was previously injected at the eight-cell stage with a nuclear localized mRNA that codes for a protein which fluoresces in both green (far left) and red (middle) when hit with blue or green light, respectively. The far right panel shows the two other panels in overlay. Embryo mounted by Mario Vargas-Vila.
Courtesy of Crystal Chaw
A flattened P.hawaiensis embryo at germ cap stage (~25h of development). One cell of this embryo was previously injected at the eight-cell stage with a nuclear localized mRNA that codes for a protein which fluoresces in both green (far left) and red (middle) when hit with blue or green light, respectively. The far right panel shows the two other panels in overlay. Embryo mounted by Mario Vargas-Vila.
Courtesy of Crystal Chaw